Nature Magazine Paper - Using Machine Learning to Predict Autism Genes
A new study from Princeton University has found some new candidate genes related to autism.
Although researchers estimate that there are hundreds of genes associated with autism, only a small fraction of them actually have clear experimental evidence that they are related to autism. A study published in the Nature Journal on August 1 aims to change the status quo by using a big data and machine learning approach to conduct a genome-wide prediction of pandemic autism disorder (ASD). Arjun Krishnan, the first author of the study, told us how their results will help early diagnosis and treatment of ASD.

Q: Can you briefly summarize your research?
Arjun Krishnan: Autism Spectrum Disorder (ASD) has a strong genetic basis and is expected to have 400-1000 related genes, but currently only about 65 autism genes have been found. Due to the complexity of ASD, photo-sequencing or genetic research alone is not enough to reveal the genetic basis of autism. Therefore, we decided to adopt a supplementary data-driven approach to solve this problem. The method we use is based on how the previously learned pattern of autism genes is linked to a specific network of genes in the human brain. We use these patterns to identify new autism genes.
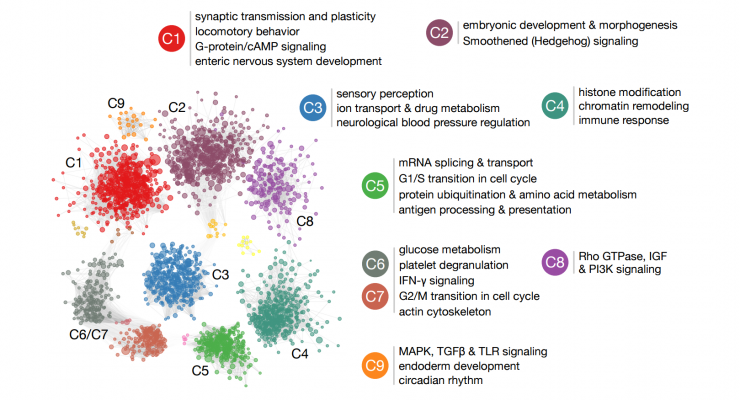
The most important result is a comprehensive supplementary prediction of autism-related genes in the genome. In other parts of the study, using these genome-wide ASD candidate genes and brain networks, we have identified the stages and regions of brain development and the specific cellular functions that may be disrupted in autistic patients. We have also set up an interactive website where any biomedical researcher or clinician can access and review the results of using our research. Â 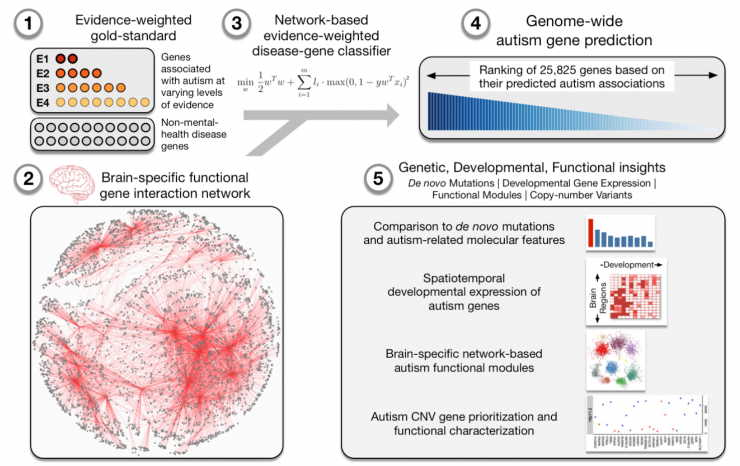
Q: What is the significance of your findings?
Arjun Krishnan: We predicted hundreds of "new" candidate genes that were never identified or involved in previous autism-related genetic studies. For geneticists, this means that they can use our predictions to perform sequencing studies directly and find autism-related genes faster and cheaper. Researchers can use them to distinguish and interpret the results of the research on ASD whole genome sequencing. Finally, biomedical researchers can use these data and related analyses to fully investigate the new autism genes and their associated functions, development, and structural effects.
Q: Can you explain to us how the machine learning method is applied to this study?
Arjun Krishnan: This technology is basically similar to the “social networking†method used by Facebook. People are related to each other in social context. Facebook first finds your friends in social networks and recommends that you add a secondary school student. Be a friend, and then recommend it through your friends in the social network.
We have established a specific brain gene network, which is a map of how the genes depend on each other in the brain. Using this network, we used similar concepts to predict new ASD genes. First, we discovered in our brain network the companions that were already associated with the ASD gene and then identified other genes in the network that were associated with these collaborating genes.
This idea, together with other ideas, formed a machine learning framework for our system predictions.
Q: What methods did you use to achieve these results?
Arjun Krishnan: What we use to do ASD gene prediction is a machine learning method that can learn how to identify autism genes that are linked to other genes in the gene network and then use these patterns to predict new ASD genes. The genetic network we use shows how genes work together in cells in the brain, or intuitively a functional map of the brain's molecular level.
We collected genes related to autism from all possible sources, including those with direct or indirect evidence, and at the same time tracked the evidence for each gene. We then established a network-evidence-weighted disease gene classifier to learn the connection patterns of known ASD genes in the brain network (taking into account the level of evidence for each gene) and then use a data-driven model to predict each gene in the genome. Potential relevance to ASD.
Q: How is this approach different from previous genetic prediction methods?
Arjun Krishnan: Our research has two major contributions to traditional genetic prediction methods. The first is that we use a genome-wide tissue-specific network. The origins and manifestations of human diseases are in specific tissues and cell types in the human body, such as hypertension-kidney, or autism-brain. Therefore, to accurately describe which genes are related to diseases that are similar to autism, we need to understand and predict what happens to these genes in the brain, rather than in parts of the body outside the brain. We achieved this result by using specific brain network genes in the human genome, which is based on the fusion of thousands of genomic experiments into specific brain signals.
The second contribution is the use of weighted evidence for classification. We have carefully planned a set of ASD-related genes from multiple sources and tracked the reliability of these sources, using their level of evidence to make new prediction methods for our machine learning. Predictions made in this way are much more accurate than predictions based on high-confidence genes.
Q: What does your study mean for ASD?
Arjun Krishnan: There is a great need for a gene or molecular test to diagnose ASDs. As early as possible during brain development, patients should be treated with drugs or other interventions based on their genetic makeup. By experimenting with these candidate genes, researchers helped researchers effectively reduce the genetic basis and genetic screening of ASD. Our results have brought them closer to these goals.
Q: What do you think is the greatest potential of machine learning in medical research?
Arjun Krishnan: The greatest potential of machine learning that I have seen is to use it to deal with this problem—accurately predicting the health and disease state of an individual’s genetic makeup. Our work is a huge step forward in the area of ​​major diseases, helping to identify the disease's "characteristic" possible definition of the disease, hoping it can be used to predict the disease. What is important is that in the process of pursuing this goal, it is not simply to conduct independent research in the field of machine learning or biomedical research. It is even more important to consider how these two fields can work together to exert great potential.
Q: How is the next step planned?
Arjun Krishnan: The next step we are thinking about is how to apply our predictions to the whole genome sequencing study of autism patients. This is very exciting to us. Research on genome-wide sequencing needs to face quite complex situations. Our predictions can help researchers focus on a variable that falls near or close to the gene and we can identify it as an option highly related to the ASD gene.
PS : This article was compiled by Lei Feng Network (search “Lei Feng Network†public number) and it was compiled without permission.
Via ResearchGate
Pcb Speaker,Pcb Subwoofer,Pcb Mount Speaker,Pcb Mounted Speaker
NINGBO SANCO ELECTRONICS CO., LTD. , https://www.sancobuzzer.com